Molecular simulation of pH (Low) Insertion Peptide pHLIP¶
The bigger picture¶
A major challenge in cancer treatment is off-target side effects, where the cancer drug attacks the healthy cells in addition to the cancerous ones. Distinguising cancerous tumors is made even harder by the heterogeneity of these tumors and rapid mutations that lead to drug resistance. A potential solution is to exploit the properties of tumor microenvironment that is universal of all cancer cells. One such property is the extracellular pH, which is lower in tumor cells (~ 6.8) than healthy tissues (~ 7.2).
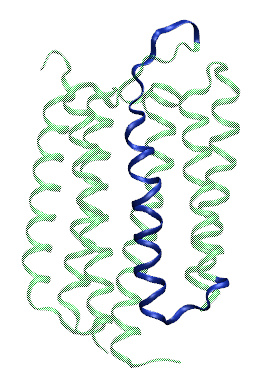
A peptide derived from the helix C of bacteriorhodopsin (see figure on left) has exactly this property, and has been named pH (Low) Insertion Peptide, pHLIP. This peptide is soluble and unstructured in solution (State I, see figure below) and binds to surface of lipid membranes under neutral and alkaline pH (State II, see figure below). However, once the pH drops to acidic level, pHLIP spontaneously forms a transmembrane helix. This property makes pHLIP a great candidate for cancer diagnostic and therapeutic. However, pHLIP insertion happens at a pH ( ~ 6.2) that is much lower than the pH of most cancerous tumors. Thus, modifications to the pHLIP sequence are necessary to develop pHLIP into a novel cancer-targeting agent. Such modifications require a detailed understanding of the properties of pHLIP at a molecular level. This is the focus of my PhD research.
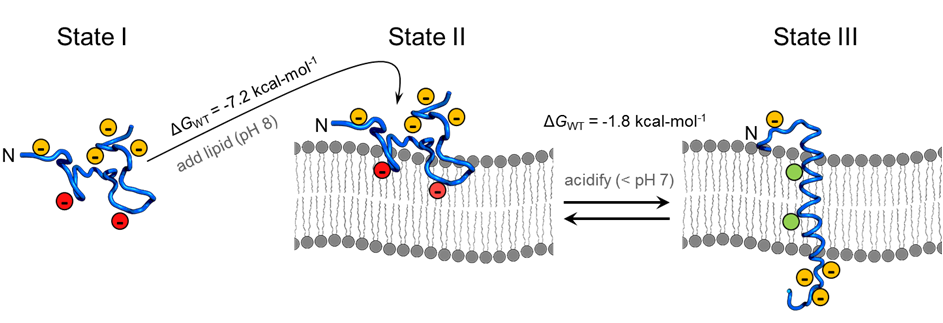
State I of pHLIP¶
While state I of pHLIP has mostly been ignored, I took a second look. This was due to two factors: (i) While conventional wisdom said pHLIP was unstructured in state II, some results showed otherwise (ii) Theories of peptide folding predict a equilibrium between completely coiled and partially folded states of a peptide that has a propensity to form a secondary structure. This rasied the question: Is there partial secondary structure in State I?
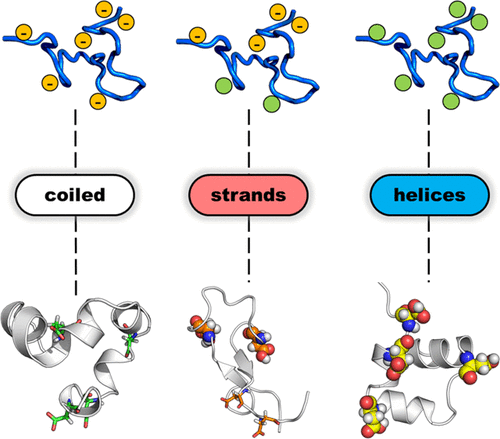
I have performed long timescale (~ 2 microsecond) simulation of pHLIP in order to answer these questions. I titrated selective acidic residues as well as introduced unnatural mutants that were shown previously to have altered pH of membrane insertion. Such long timescales became feasible by using GPU-accelerated Amber simulation engine in combination with implicit solvation, which bypasses the need to explicitly define the solvent atoms thereby significantly reducing the computation time. This study has elucidated the influence of site-specific titration and mutation on the helix-forming propensity of pHLIP. Moreover, we have discovered transient sampling of the ramachandran space of pHLIP that was not known heretofore. Click here to read our publication.
While this is a good starting point, we eventually want to understand the interactions of pHLIP with water molecule. I have achieved this by the use of gaussian accelerated molecular dynamics (GaMD). This work is currently under progress.
State II of pHLIP¶
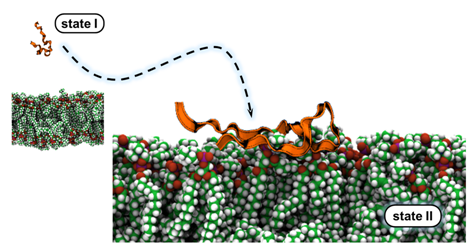
Most of the magic really begins in state II. Unfortunately, very little is known about conformation of pHLIP in state II and its orientation with respect to the bilayer normal. I have performed long timescale ( ~ 800 ns) simulation of pHLIP placed on a model lipid membrane at various orientations. This preliminary study has revealed the complex interplay of various non bonded interactions between pHLIP and the POPC bilayer that govern membrane-binding of pHLIP. We have observed desolvation of pHLIP and burial of one arginine and two tryptophan residues into the lipid bilayer, each of which is consistent with experimental results. Additionally, this study has provided an atomistic view of the favorable binding orientation of pHLIP. Until now, all that was known about this orientation are distances of the C-beta carbons of three alanine residues from the phosphate headgroups. We also found partial helix formation in surface-bound pHLIP. Click here to read our publication.
Effect of salt concentration¶
While it is known that pHLIP aggregates under higher salt concentration, little is known until now about the influence of ionic strength on lipid-binding. Uncovering this effect is essential for administering pHLIP at biological salt concentrations. I am performing molecular dynamics simulation of pHLIP-lipid system with varying ionic strength to gain a molecular-level understanding of the role played by the ions. We have collaborated with the group of [Prof. Francisco Barrera](https://barreralab.com/) for this work. The Barrera group has a long tradition of working with pHLIP and pHLIP-like peptides, including the influence of bilayer elctrostatics (see below) and ionic strength on pHLIP behavior. Together, we have revealed the specific region(s) of pHLIP which see the influence of salt concentration and how that affects pHLIP's affinity for protonation. Click here to read our publication.
Effect of bilayer electrostatics¶
Influence of anionic lipid headgroups on pHLIP-binding has mostly been overlooked until now. Most cancerous tumors have higher levels of POPS lipid in the outer leaflet, which can be expected to repel the negatively charged pHLIP. Some recent experiments, including experiments from the Barrera group (see above), have began shedding light on this issue. I have performed replica exchange with solute tempering (REST2) simulation of pHLIP-lipid system to gain a fundamental understanding of the interactions between pHLIP and the lipid bilayer. In my study, I have systematically varied the POPS and POPE content of the lipid membrane. My simulations have helped explain some of the discrepancies of the experimental results. I have used XSEDE's resources for this study. I would also thank Wei Jiang for providing me a pre-release version of the NAMD 2.12 implentation of REST2. This not only helped me conduct this work in a timely fashion, but also gave me hands-on experience in building NAMD on XSEDE's resource. This work is currently being prepared for submission.